From GWAS to signal validation: A novel method for estimating SNP genetic effects while preserving genomic context
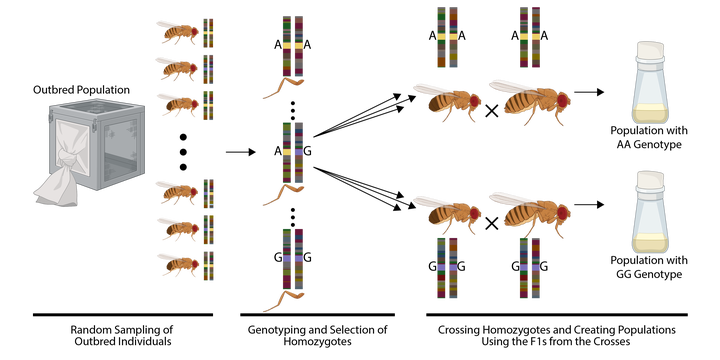
Abstract
Validating associations between genotypic and phenotypic variation remains a challenge, despite advancements in association studies. Common approaches for signal validation rely on gene-level perturbations, such as loss-of-function mutations or RNAi, which can only identify effects coarsely. More recent CRISPR-based methods can validate associations at the SNP level, but have significant drawbacks, including resulting off-target effects and being both time-consuming and expensive. Both approaches usually modify the genome of a single genetic background, limiting the generalizability of experiments. To address these challenges, we present a simple, low-cost experimental scheme for validating genetic associations at the SNP level in outbred populations. The approach involves genotyping live outbred individuals at a focal SNP, crossing homozygous individuals with the same genotype at that locus, and contrasting phenotypes across resulting synthetic outbred populations. We tested this method in Drosophila melanogaster, measuring the phenotypic effects of a polymorphism at a naturally-segregating cis-eQTL for the midway gene. Our results demonstrate this method’s utility in SNP-level validations of naturally occurring genetic variation regulating complex traits. This method provides a bridge between statistical discovery and validation in the natural context of outbred populations, offering a potential solution to the limitations of current methods.